einstein (São Paulo). 17/Dec/2025;24:eAO0757.
Characterization of a novel MSH2 variant in Lynch syndrome: clinical data and complementary bioinformatics assessment
DOI: 10.31744/einstein_journal/2026AO0757
Highlights
■ Family identified with recurrent colorectal cancer, suggestive of Lynch syndrome.
■ A new variant was detected, NM_000251.3(MSH2):c.1894_1898del (p.Ile633Lysfs*9).
■ The variant causes a frameshift mutation leading to a premature stop codon, suggesting pathogenicity.
■ Using bioinformatics tools, we investigated possible escape mechanisms, such as the presence of isoforms without the mutated exon.
■ Location of the variant suggests that it causes non-sense mediated decay, which is reinforced by immunohistochemistry findings showing absence of MSH2 protein expression.
■ Detailed assessment allows this variant to be classified as probably pathogenic/pathogenic.
ABSTRACT
Objective:
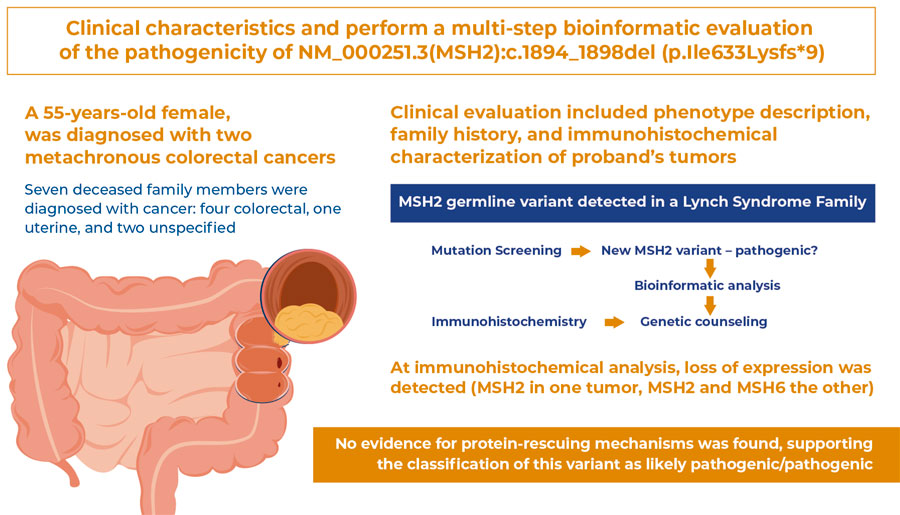
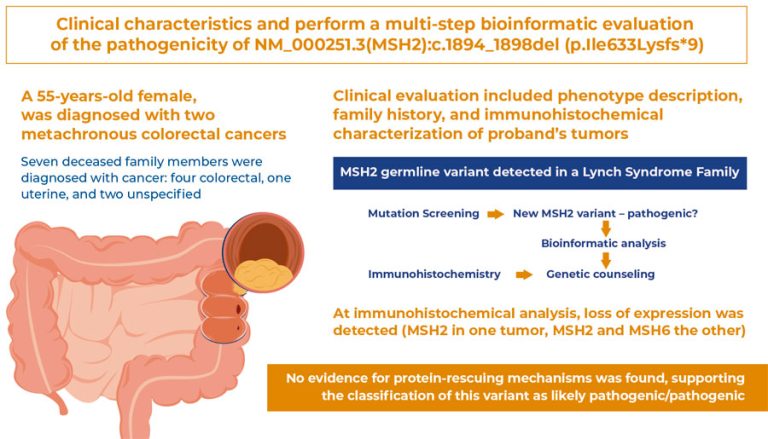
To describe the clinical characteristics and perform a multi-step bioinformatics evaluation of the pathogenicity of NM_000251.3(MSH2):c.1894_1898del (p.Ile633Lysfs*9), an MSH2 germline variant detected in a family with Lynch syndrome.
Methods:
Clinical evaluation included description of phenotype, family history, and immunohistochemical characterization of the proband’s tumors. For pathogenicity classification according to the American College of Genetics and Genomics/Association for Molecular Pathology (ACMG/APA) criteria, bioinformatics analyses included: (i) literature and database screening, searching for the variant allele frequency, case reports, or functional studies, including ClinVar, VarSome, Ensembl, PubMed, EVA, and ABraOM; (ii) prediction of variant impacts using ExPASy Translate, Pfam, and Modeller 9.24; and, (iii) mechanisms that could mitigate the effects of the variant included alternative splicing and exon skipping (UniProt and GTex) and nonsense-mediated decay (NMD; MutationTaster2021).
Results:
The proband, a 55-year-old female, was diagnosed with two metachronous colorectal cancers. Immunohistochemical analysis showed loss of expression ( MSH2 in one tumor, and MSH2 and MSH6 in the other). Seven deceased family members were diagnosed with cancer (four colorectal, one uterine, and two unspecified). This variant caused a stop codon in MSH2 exon 12 of 16. When translated, the protein loses 294 C-terminal residues, which may prompt protein degradation. If the mutated protein escapes degradation, dimerization and DNA-binding domains will be present. Therefore, negative dominance effects were possible. No isoforms ending in exon 12 have been identified in the literature or in RNA splicing databases. A stop codon before the last exon-exon boundary indicated the occurrence of NMD.
Conclusion:
No evidence of protein-rescuing mechanisms was found, supporting the classification of this variant as likely pathogenic/pathogenic.
[…]
107