07/Apr/2025
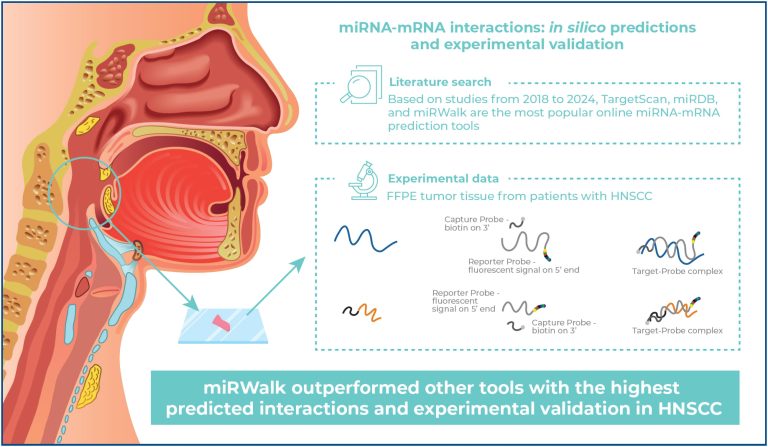
Comparative analysis of miRNA-mRNA interaction prediction tools based on experimental head and neck cancer data
DOI: 10.31744/einstein_journal/2025AO1372
Highlights ■ miRWalk had the highest predicted interactions and validated miRNA networks in HNSCC. ■ Around 3.3% of interactions overlapped across tools, emphasizing the need for multitool approaches. ■ Dysregulated genes and miRNAs were tied to cancerdriving PI3K-Akt and Wnt pathways. ■ The validated approach highlights the importance of integrating computational and molecular data. ABSTRACT Objective: Head and neck squamous cell carcinoma (HNSCC) has a poor prognosis largely due to late diagnosis and a lack of reliable biomarkers. MicroRNAs (miRNAs), […]
Keywords: Algorithms; Biomarkers; Computational biology; Gene expression; MicroRNAs; Squamous cell carcinoma of head and neck; Target prediction tool
02/Oct/2020
In silico investigation of heparanase-correlated genes in breast cancer subtypes
DOI: 10.31744/einstein_journal/2020AO5447
ABSTRACT Objective To investigate the possible genes that may be related to the mechanisms that modulate heparanase-1. Methods The analysis was conducted at Universidade Federal de São Paulo, on the data provided by: The Cancer Genome Atlas, University of California Santa Cruz Genome Browser, Kyoto Encyclopedia of Genes and Genomes Pathway Database, Database for Annotation, Visualization and Integrated Discovery Bioinformatics Database and the softwares cBioPortal and Ingenuity Pathway Analysis. Results Using messenger RNA expression pattern of different molecular subtypes of […]
Keywords: Breast neoplasms; Computational biology; Heparanase-1