07/abr/2025
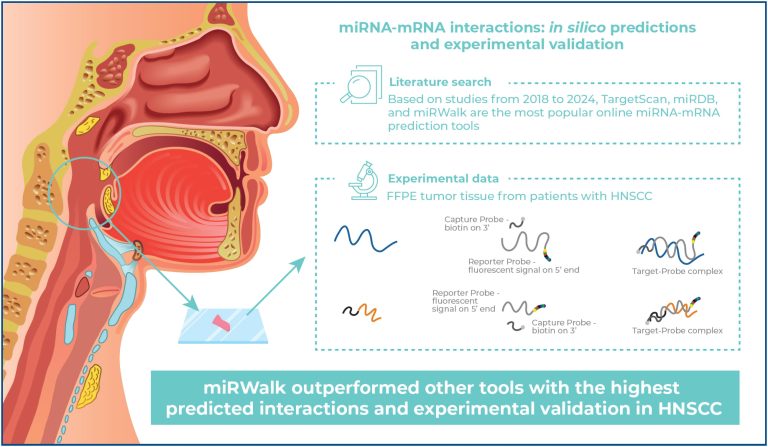
Comparative analysis of miRNA-mRNA interaction prediction tools based on experimental head and neck cancer data
DOI: 10.31744/einstein_journal/2025AO1372
Highlights ■ miRWalk had the highest predicted interactions and validated miRNA networks in HNSCC. ■ Around 3.3% of interactions overlapped across tools, emphasizing the need for multitool approaches. ■ Dysregulated genes and miRNAs were tied to cancerdriving PI3K-Akt and Wnt pathways. ■ The validated approach highlights the importance of integrating computational and molecular data. ABSTRACT Objective: Head and neck squamous cell carcinoma (HNSCC) has a poor prognosis largely due to late diagnosis and a lack of reliable biomarkers. MicroRNAs (miRNAs), […]
Palavras-chave: Algorithms; Biomarcadores; Biologia computacional; Expressão gênica; MicroRNAs; Squamous cell carcinoma of head and neck; Target prediction tool
02/out/2020
Investigação in silico dos genes correlacionados à heparanase nos diferentes subtipos de câncer de mama
DOI: 10.31744/einstein_journal/2020AO5447
RESUMO Objetivo Investigar os genes que podem estar relacionados aos mecanismos que modulam a heparanase-1. Métodos A análise foi realizada na Universidade Federal de São Paulo, utilizando dados fornecidos por: The Cancer Genome Atlas, University of California Santa Cruz Genome Browser, Kyoto Encyclopedia of Genes and Genomes Pathway Database, Database for Annotation, Visualization and Integrated Discovery Bioinformatics Database e os softwares cBioPortal e Ingenuity Pathway Analysis. Resultados Usando o perfil de expressão de RNA mensageiro de diferentes subtipos moleculares de […]
Palavras-chave: Biologia computacional; Heparanase; Neoplasias da mama